Applications of DNA Foundation Models, part 2
Genome-scale generation, patient selection strategies, multiple variant effect prediction, SaaS meets DNA FMs
This is the second and much shorter part of the series. Part 1 can be found here. This post looks at four more applications of DNA Foundation Models (DNA FMs) I find interesting. Some are relatively obvious but obvious applications should be considered too.
Engineering genome-scale drug manufacturing systems for biologics and crops
Developing biomarkers for disease detection, patient selection strategies for clinical trials, and monitoring response to treatment
Multiple in silico mutagenesis prediction
DNA FMs As-A-Service
1. Engineering genome-scale drug manufacturing systems for biologics and crops
Look no further than Brian Hie and team’s work fine tuning Evo2 (this is a gross oversimplification, please read their technical blog or preprint - it’s fantastic) to generate new bacteriophages that are functional when synthesized and have the ability to do things like overcome and kill E. Coli that are phage-resistant. A new era of genome design has started. Controllable generation of genomes with unique and specific functions is possible.
To flesh this idea out a little more, consider the problem of designing a 100 base pair sequence with a specific property defined by a known objective function. Suppose a company has collected data on their problem of interest and has an oracle prediction model. How do we search the 4100 possible sequences? If each generate-predict loop takes 1 nanosecond, naively searching the space will take 1043 years. Harnessing DNA FMs like Evo2 to generate sequences that reflect natural sequence variation and fitness allow us to arrive at optimal solutions faster. Further, we can take DNA FMs and fine-tune prediction heads using proprietary data on the optimization parameters of interest (e.g., antibody yield, drought resistance, etc.) to develop accurate and generalizable solutions.
Asimov has developed CHO cell line data and simulation models for antibody optimization. Ginkgo too. Agricultural companies have crop-related data and leverage predictive models to guide which edits may be most effective (Benson Hill, Ohalo, Inari, Corteva, Cropwise, Pairwise, Tropic). It is unclear if any of these companies are currently using DNA FMs.
2. Developing biomarkers for disease detection, patient selection strategies for clinical trials, and monitoring response to treatment
DNA FMs are a new tool for discovering genomic features that can aid in early detection of diseases like cancer and neurodegenerative diseases, monitoring response to treatment, and identifying patients who will respond to a therapy. DNA FMs can contribute to developing new low-coverage WGS features that capture transcription factor binding sites and other transcriptome- or epigenomic information from cell-free DNA. The devil is in the details here. Implementing this technology in this space and proving its value will be difficult. I think clinical trial patient selection or switching patients from one therapy to another are probably the first two places where DNA FMs could be leveraged.
Spotlight on Polygenic Risk Scores. Polygenic risk scores (PRS) are used by embryo selection companies (e.g., Orchid Health), clinical risk and prevention companies (e.g., Allelica). PRS are actively being developed by many academic groups. Pradeep Natarajan and Nilanjan Chaterjee are academic leaders in the PRS cardiology and oncology space, respectively. PRS are based on estimating effect sizes of each variant in the set and using that as a feature to predict risk or the desired outcome. The fraction of variants selected that are causal to the disease determines the performance of the PRS. We can enrich for biologically meaningful variants by leveraging DNA FMs.
3. Multiple in silico mutagenesis prediction
How can we predict the effect of multiple distant variants across tissues and genomic tracks? Suppose you want to understand the effects of 5 different mutations and a translocation on RNA expression, cell state, and methylation patterns. Currently, we can’t do this. One way to solve this problem is to scale context length.
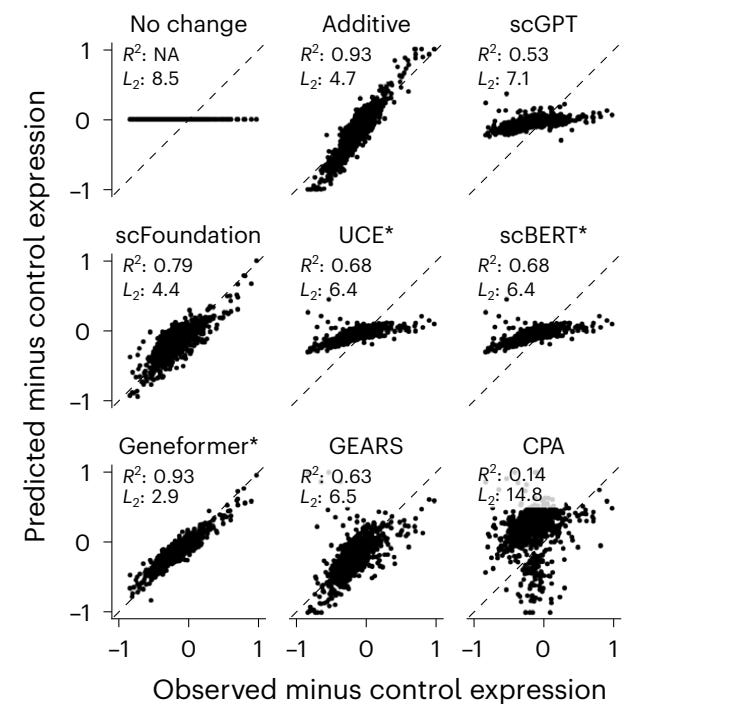
Currently, we rely on large-scale CRISPRi screens of combinations of variants with multi-omic reporting. But, this method quickly becomes very costly and combinatorically infeasible. We may be able to adapt DNA FMs to do this in silico. There are a few technical modeling questions that need to be solve to achieve this milestone. Consider testing the effect of two mutations x and y. The DNA FM prediction can be represented by some arbitrary f(.). If the mutations act independently then f(x,y) = f(x) + f(y). However, mutations need not act independently and often do not if they act on the same pathway or network. We must model their joint distribution. Simple additive models perform pretty well and much better than current neural networks, so we have a long way to go.
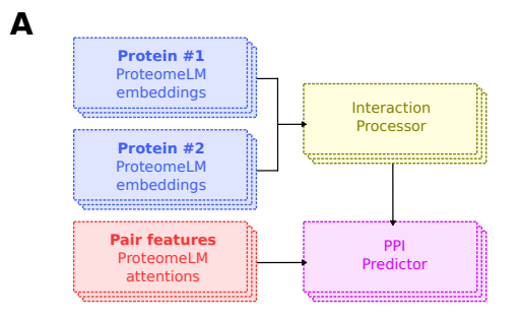
One strategy could be to borrow from ProteomeLM’s method (see below figure) for modeling protein-protein interactions by introducing pair attention features and an interaction process (see schematic from ProteomeLM paper). We also need to capture pathway information given that two mutations in the same pathway (e.g., KRAS and MAPK) can cancel or amplify one another beyond what would be expected under additivity.
The follow-on effect of doing this is being able to propose specific edits to achieve a given cellular outcome. For example, we could predict lethal interactions of higher order combinations and greatly reduce preclinical costs of drug development.
4. DNA FMs As-A-Service
SaaS meets DNA FMs. Basically, serve different prediction (e.g., Borzoi, AlphaGenome, Yorzoi) and generation models (e.g., DFM, diffusion, autoregression-based models) with linkage between the two. On top of that, integrate a front-end LLM and optimization schema to generate guided sequences. Tamarind Bio offers a similar service for protein models. We can also offer fine-tuning of the models we serve for client-specific applications given a custom dataset. LatchBio offers a suite of bioinformatic solutions (AlphaFold, BLAST, Cell Ranger, RFDiffusion) including DNAChisel (simple search and optimizer) and could expand to offer DNA FMs too.